Bioinformatics of Protein Structure is a module on the online MSc in Bioinformatics run jointly by the University of Leeds and the University of Manchester.
One of the activities on this module involves a group of students performing a phylogenetic analysis on a family of proteins. In other words, they have to deduce a "family tree". First they have to search the public protein sequence databases to find the protein family members. The whole activity spreads over 6-7 weeks.
One of the aspects of the activity that provided cause for concern was the need for what was termed a scriba, a clerk who would maintain an official list of all the sequences that the group had found. The job of scriba was really too big a burden to place on a group member especially as the number of sequences was growing rapidly each year.
It was obvious that some sort of electronic scriba was required backed up by a searchable database of protein sequences.
The author had been pootering about ineffectually for several months trying to write a suitable Web application. When the SOCKET project matured, the decision was made to use this and, lo-and-behold, in about 4 or 5 evenings work, the project had been completed. The Bodington VLE that was used to deliver the module now had a new resource: Protein Sequence Repository.
Below is a brief description of how the new Web service was created.
The basic object to be stored in the MySQL database is a protein Sequence which has the following fields:
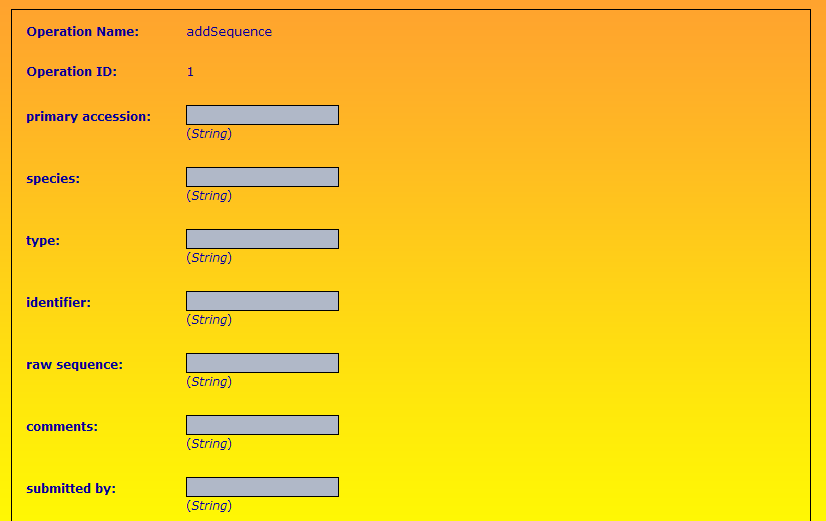
primaryAccession (String): a unique identifier
species (String): species to which the protein sequence belongs eg Homo sapiens
type (String): abbreviation representing the type of protein eg RBP for retinol binding protein
identifer (String): a unique identifer created from the species and type
rawSequence (String): the protein sequence eg MHFPYYGCG...
comments (String): general remarks about the sequence
submittedBy (String): submitter's initials
timeOfCreation (String)
lastEdited (String)
A database in MySQL was created with the fields identified above.
The new NetBeans 5.5 IDE had just come out of beta and this seemed like a good opportunity to put it through its paces.
A class on which the Web service was to be based was created: LipocalinsService.java. Lipocalins were the class of proteins used in the original activity.
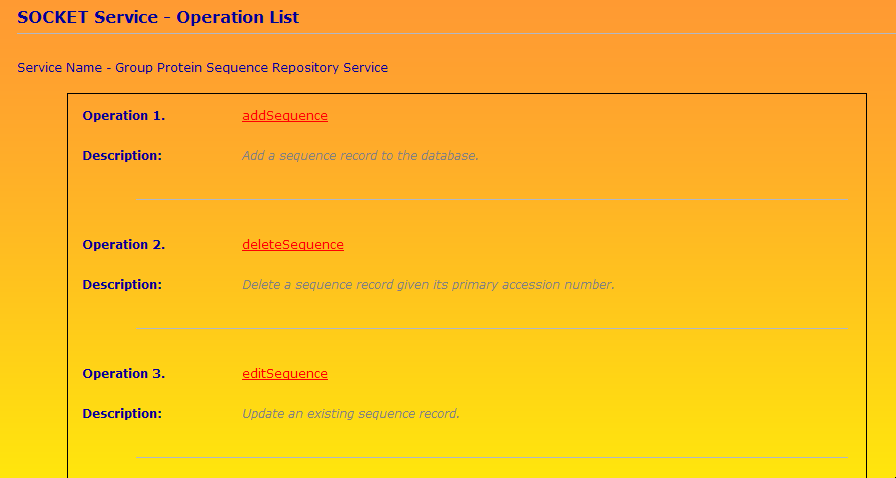
A facade pattern works well for quickly putting together an RPC (Remote Procedure Call) style Web service. The LipocalinFacade class has thirteen methods corresponding to the operations in Web service: addSequence, deleteSequence, editSequence, and so on.
Using the facade pattern the methods can be implemented in any fashion - using flat files or a relational database, for example. The code can be refactored and tinkererd with without having to change the WSDL or the SOCKET client.
In order to make each of the methods in LipocalinsService into a Web service operation, Java annotations are used. An example is given below for the method that returns all sequence records.
@WebMethod(operationName="GetAllSequenceRecords")
public String getAllSequenceRecords()
{
return lipocalinFacade.getAllSequenceRecords();
}
A bit of crude plumbing produced some SQL that performs the 13 Web service tasks.
Press the NetBeans "build" button and, wallop, there's your service all packaged up in a nice war file ready to drop into Tomcat.
The easiest way to socketize the Web service is to get the WSDL document by appending "?wsdl" to the URL of the live service.
This was the nice document/literal WSDL document that was returned: LipocalinsService.wsdl (I've edited out the address of the server on the portType).
Whack that through Saturn and the SOCKET client war file is generated: call this lipocalin_service.war.
Some customisation of the SOCKET stylesheets and that's that.
A screenshot of part of the operation list is shown below.

Part of the input form for addSequence is shown below.
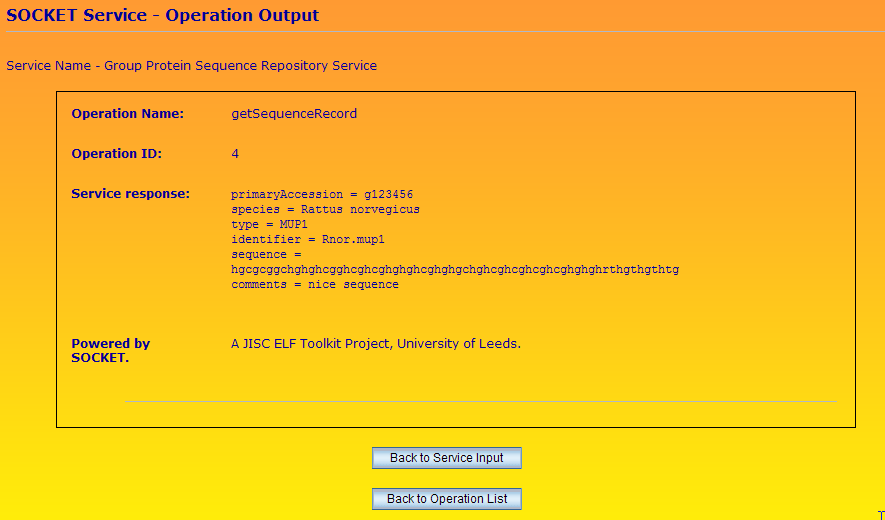
And finally, an example of the result of a getSequenceRecord request is given.
The service produced can simply be enjoyed as a loose application and client pair.
In order to make the service available as a resource in Bodington, the socket2bod.jar file must be prepared.
There are details about how this can be done on the SOCKET site.
I think the code might eventually find its way onto our CVS code repository - there are many ways in which it might be improved. In the meantime, if anyone wants to crank this up on their own machinery, contact me at b.p.clark@leeds.ac.uk.