Gel filtration
A wide range of biological molecules can be separated on the basis of differences in their size and shape which lead to differences in their ability to penetrate porous matrices. This procedure is known as molecular sieve chromatography or molecular exclusion chromatography. A variety of porous matrices can be employed depending on the nature of the molecules to be fractionated. For protein purification purposes, the matrix typically consists of porous beads of an inert, highly hydrated gel and so the process is often referred to as gel filtration.
Common commercial gel matrices are Sephadex (dextran beads), Sepharose and Bio-Gel A (agarose) and Bio-Gel P (polyacrylamide), whilst other materials such as polyacryloylmorpholine and various polystyrenes have also been used. The dextran, agarose and polyacrylamide gel beads can all be manufactured with different degrees of porosity and so will fractionate different size ranges of proteins. The Sephadex dextran gels and Bio-Gel P polyacrylamide gels allow fractionation of globular proteins up to about 800,000 molecular weight whereas agarose gels, because of their greater porosity, can separate protein molecules and macromolecular complexes up to several million molecular weight. More recently, cross-linked dextran gels (Sephacryl) have been marketed which are exceptionally rigid, stable matrices which can fractionate proteins of molecular weights up to 8 million. The rigidity and stability of polyacrylamide gels can be improved by including agarose in the gel matrix. Polyacrylamide/agarose gels of this type are also available commercially (Ultrogel). New types of commercial gel are constantly being developed. The table below summarizes the properties of the matrices used in this program.
Characteristics of gel filtration media used in this simulation.
|
|
|
Approximate fractionation range for peptides and globular proteins (molecular weight) |
|
Sephadex G-50 ¹ |
dextran |
1500 - 30000 |
|
Sephadex G-100 ¹ |
dextran |
4000 - 150000 |
|
Sephacryl S-200 HR ¹ |
dextran |
5000 - 250000 |
|
Ultrogel AcA 54 ² |
polyacrylamide/agarose |
6000 - 70000 |
|
Ultrogel AcA 44 ² |
polyacrylamide/agarose |
12000 - 130000 |
|
Ultrogel AcA 34 ² |
polyacrylamide/agarose |
20000 - 400000 |
|
Bio-Gel P-60 ³ |
polyacrylamide |
3000 - 60000 |
|
Bio Gel P-150 ³ |
polyacrylamide |
15000 - 150000 |
|
Bio-Gel P-300 ³ |
polyacrylamide |
60000 - 400000 |
¹
Sephadex is a registered trademark of Pharmacia PL.Fractionation of a protein mixture by gel filtration is typically carried out as follows. The gel beads, present as a slurry in the chosen buffer, are poured into a glass or plastic chromatography column of suitable dimensions and allowed to settle by gravity. After washing and equilibration of the column with buffer alone, the protein mixture in buffer is applied to the top of the column and the eluate is collected at the column base in a series of fractions. As the proteins pass down the column they penetrate the pores of the gel beads to different extents and so travel down the column at different rates. All proteins which exceed the maximum size of the pores will be unable to enter the beads. These proteins will therefore distribute only in the solution between the beads and elute from the column first in the so-called 'exclusion volume' (or void volume). All proteins below the minimum size of the pores will equilibrate completely with the buffer inside and outside the gel beads and so spend a proportion of their time inside the beads. These proteins will therefore move more slowly through the column and will be eluted last, together with any other small molecules present in the original protein mixture such as salt ions. The total volume of the column is called the bed volume. Therefore these proteins will elute in a volume of buffer very close to the bed volume of the column (although somewhat less due to the volume physically occupied by the gel matrix itself). The pores in the beads are not of exactly identical sizes but rather span a narrow range of sizes. Some proteins will have sizes very similar to the size range of the pores and so be excluded from some pores whilst entering others. These proteins of intermediate size will therefore be partially excluded from the beads to an extent that depends on their size and shape. They will elute from the column in order of molecular weight with the largest proteins eluting first and the smaller proteins later.
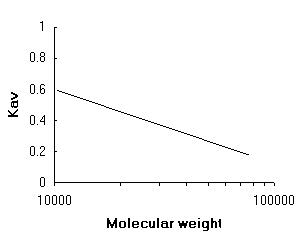
Careful choice of gel type is important for the optimal fractionation of complex protein mixtures. Use of an inappropriate gel type may lead to the protein of interest either being totally excluded from the gel beads and so eluting with many other proteins in the void volume or, conversely, it may be able to equilibrate with the whole gel volume and so elute with many other proteins in the salt fraction near the bed volume. In both situations the resolution of the fractionation is poor. Ideally the gel pore size should be such that the desired protein is partially excluded from the gel beads, a condition which leads to the greatest degree of fractionation from other protein species. For partially excluded proteins, a plot of log(protein molecular weight) versus Kav, leads to a straight line relationship, where
|
Kav |
= |
elution volume - void volume total volume of the packed matrix - void volume |
Linear relationship between Kav and log(molecular weight)

Thus a column can be calibrated with standard proteins of known molecular weight, noting their elution volumes, and these data used to draw such a graph. Subsequently, during fractionation of the sample protein mixture on the same column, measurement of the elution volume of the protein of interest allows its molecular weight to be deduced by reference to the standard curve. However, it should be noted that the shape of protein molecules also plays an important role in gel filtration. Long, extended polypeptides and proteins tend to behave as though they were larger, globular protein molecules. Therefore a calibration curve is only as accurate as the nature of the protein standards used to construct it will allow.
When first applying gel filtration for any particular protein purification, it is good practice to fractionate the protein mixture on several gel matrices of different porosity, collect fractions and assay each for protein content and enzyme activity. Empirically one can then choose a matrix that gives good resolution of the desired protein from other proteins whilst also yielding some information about its molecular weight - information which may prove extremely useful in subsequent purification steps.